Here are a couple tips based on my experience using SMC to complete the Coursera course, Introduction to Data Science in Python. This course is the first installment of a new 5-part Applied Data Science with Python Specialization from the University of Michigan.
Examples and study assignments for the course are offered as Python3 Jupyter notebooks, i.e. .ipynb files. Students may use Coursera-hosted jupyter notebooks or any other platform that allows them to run the code. Homework is submitted by uploading a .ipynb file for each programming assignment.
The following might be helpful for students taking the course and using SMC:
1. Set Jupyter kernel.
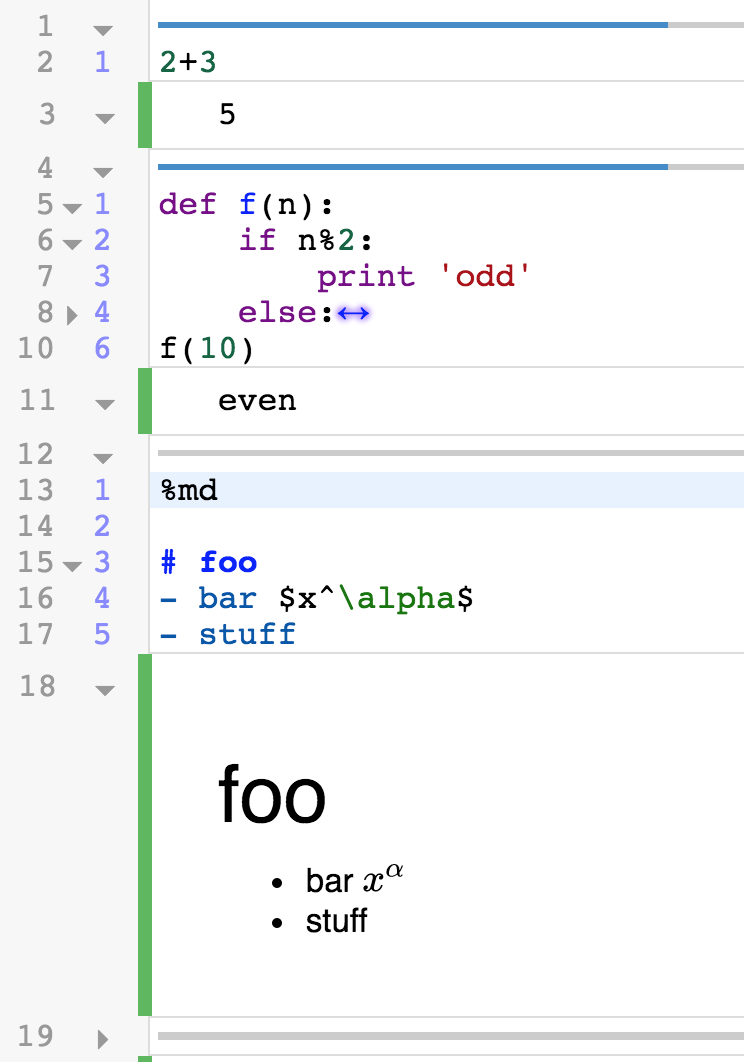
After uploading a course .ipynb file, change the kernel from Python3 to Anaconda (Python3) as shown below. This will prevent errors such as AttributeError: ‘Index’ object has no attribute ‘str’ due to different versions of pandas.

2. Convert Jupyter notebooks to Sage worksheets.
If you would rather code in a Sage worksheet than a Jupyter notebook, use the SMC script smc-ipynb2sagews to convert the files. Open a terminal file, for example mooc.term, and enter the following commands
$ cp Assignment\ 2.ipynb assgn2.ipynb
$ smc-ipynb2sagews assgn2.ipynb
/usr/local/bin/smc-ipynb2sagews: Creating SageMathCloud worksheet 'assgn2.sagews'
$ open assgn2.sagews